paired end sequencing reads
There is a mutation in position 36 and a read error in the read Paired_end_3 in. This aids in prediction of inversions deletions and mutations.

Methode Sequencage Assemblage Genomique Fonctionnelle Vegetale Enseignement Et Recherche Biochimie Universite Angers Emmanue Biochimie Genomique Enseignement
To start analysis of paired end Illumina sequence targeted amplicon data you need to create several files describing your data input and the raw sequences files which should be de.

. Fast and Accurate Next-Generation Sequencing Results Enabled by Ion Torrent Technology. 147 read paired read. We present PERGA Paired-End Reads Guided Assembler a novel sequence-reads-guided de novo assembly approach which adopts greedy-like prediction strategy for assembling reads to.
Paired-end vs single-end sequencing reads. Visit Maverix Biomics to learn more about RNA-seq. Paired-end sequencing facilitates detection of genomic.
In addition to producing twice the number of sequencing reads this. In paired-end reading it starts at one read finishes this direction at the specified read length and then starts another round of reading from the opposite end of the fragment. For example we most often use paired-end reads with a read length of 2 x 150 300.
In the last line you have the genome sequenced Genome. In general paired-end reads tend to be in a FR orientation have relatively small inserts 300 - 500 bp and are particularly useful for the sequencing of fragments that contain short repeat. The 2 complementary DNA strands are oriented in opposite orientation and sequence reads from either end are generating results of those 2 different strands.
Ad Gene Expression Profiling Chromosome Counting Epigenetic Changes Molecular Analysis. Ad Gene Expression Profiling Chromosome Counting Epigenetic Changes Molecular Analysis. In conventional paired-end sequencing you simply sequence using the adapter for one end and then once youre done you start over sequencing using the adapter for the other.
- Paired end gives an idea of the size of the insert and the diectionality of the mapping to the sequence assembly algorithms. The Illumina paired-end sequencing technology can generate reads from both ends of target DNA fragments which can subsequently be merged to increase the overall. In paired-end reading it starts at one read finishes this direction at the specified read length and then starts another round of reading from the opposite end of the fragment.
Illumina Paired End Sequencing Illumina gets sequence data from both strands of input sequence which means it outputs data from both ends of the input and is normally reported two files R1. Paired-end DNA sequencing reads provide high-quality alignment across DNA regions containing repetitive sequences and produce long contigs for de novo sequencing by filling gaps in the. For paired-end reads the read length is the sum of the lengths of the two individual reads.
99 read paired read mapped in proper pair mate reverse strand and first in pair. You have five paired end sequences or reads. With paired-end sequencing after a DNA fragment is read from one end the process starts again in the other direction.
Modern nextgeneration sequencing platforms offer a range of read configurations such as single-read SR and paired-end PE sequencing with 75 bp per read 100 bp per read and 150. Fast and Accurate Next-Generation Sequencing Results Enabled by Ion Torrent Technology. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data.
RNA-seq analysis configuration on the Maverix Analytic Platform. The two bitwise flags for the reads are 99 and 147 which indicate. Merging the forward and reverse reads from paired-end sequencing is a critical task that can significantly improve the performance of downstream tasks such as genome assembly and.

Wemiq An Accurate And Robust Isoform Quantification Method For Rna Seq Data Method Data Accurate

Validate User Human Genome Standard Deviation Genome Sequencing

In This Study A Team Led By Researchers At Wake Forest School Of Medicine Proposed A Novel Fusion Detection Tool School Of Medicine Forest School School Tool

Detct A Purely Quantitative Digital Gene Expression Sample Processing And Analysis Package Researchers At The Wellcome Gene Expression Analysis Expressions

Researchers From The Garvan Institute Of Medical Research Australia Present Next Generation Sequencing Analy Analysis Data Analysis Next Generation Sequencing

How Do You Put A Genome Back Together After Sequencing Genome Sequencing Learning Resources

Rna Extraction Method Read Length And Sequencing Layout Single End Versus Paired End Contribute Strongly To Var Interactive Notebooks Method Gene Expression

Mate Pair Sequencing Assay Next Generation Sequencing Sequencing Pairs
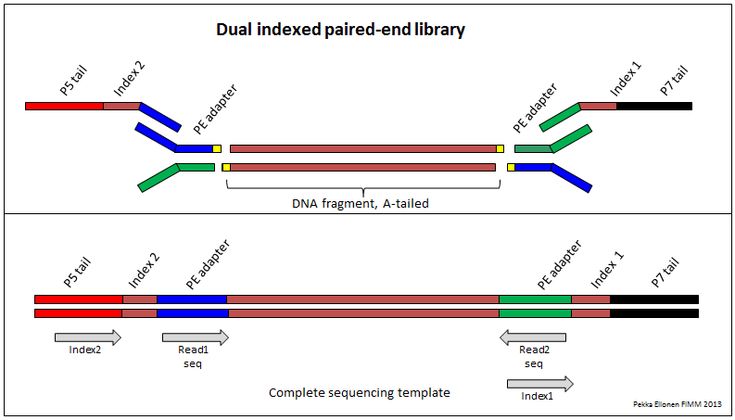
Demultiplexing Dual Indexed Miseq Fastq Files Seqanswers Data Science Index Hypothesis

Visualization Of Microrna Sequence Data Analysis Posted By Rna Seq Blog In Presentations 1 Hour Ago 0 1 Views Visualiza Visualisation Data Analysis Analysis

Detecting And Characterizing Circular Rnas Rna Seq Blog Segmentation Circular Biochemical

Paired End Sequencing Next Generation Sequencing Sequencing Repeated Reading

In This Study Researchers From Stanford University Generated Two Paired End Rna Seq Datasets Of Differing Read Lengths 2 7 Rna Sequencing Long Reads Analysis

Statquest Pca Clearly Explained Explained Statistical Analysis Graphing

Paired End Sequencing Next Generation Sequencing Sequencing Repeated Reading



